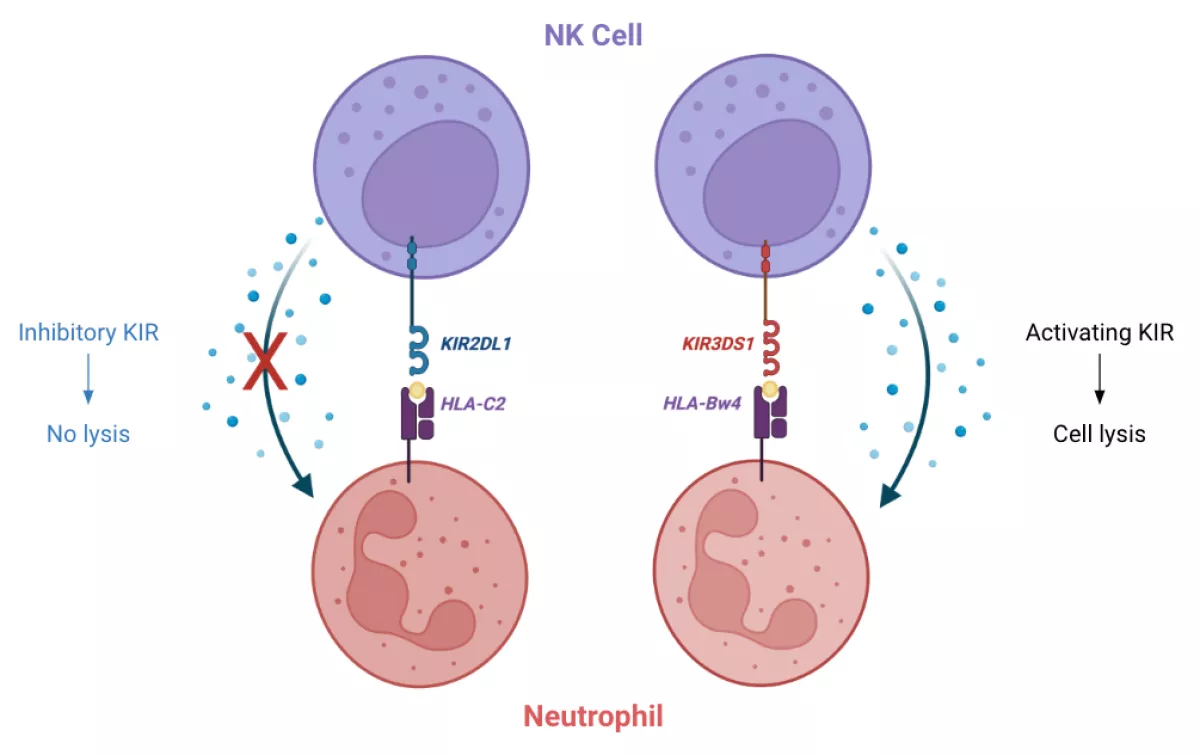
Multiple Sclerosis is an immune-mediated neurodegenerative disorder whose disease mechanism is believed to be a combination of genetic, environmental and cellular risk factors. The most prevalent genetic risk factor is the presence of the HLA-DRB1*15:01 allele, and a recently identified, prominent environmental risk factor is Epstein Barr Virus. There is also increasing evidence that natural killer (NK) cells may play a role in disease as impaired function of NK cells impacts its ability to control viral infections and perform immune surveillance. NK cell functionality is dependent upon cell-cell interactions through their cell surface receptors, the most polymorphic of which are encoded by the killer cell immunoglobulin-like receptor (KIR) genes. The KIR complex located on chromosome 19 is made up of 15 activating and inhibitory genes, each having varying degrees of sequence similarity, allelic diversity, and haplotypic variation. They facilitate NK cell functionality through their interactions with HLA class I molecules on surrounding cells. Various studies have suggested that the polymorphic nature of both KIR and HLA class I makes them potential susceptibility factors for infections and autoimmune disorders. In addition, there have also been studies showing that certain KIR-HLA pairs have been associated with protective effects against MS risk, likely due to the balance of inhibitory and activating signals. For this reason, we hypothesize that the genetic variability of KIR and their HLA class I ligands likely plays a role in risk for and/or protection against multiple sclerosis. The goal of this project is to characterize the genetic variation of KIR and HLA in the context of multiple sclerosis using existing and novel bioinformatic methods. In doing this we hope that our findings may pave the way for NK-based therapies for multiple sclerosis.